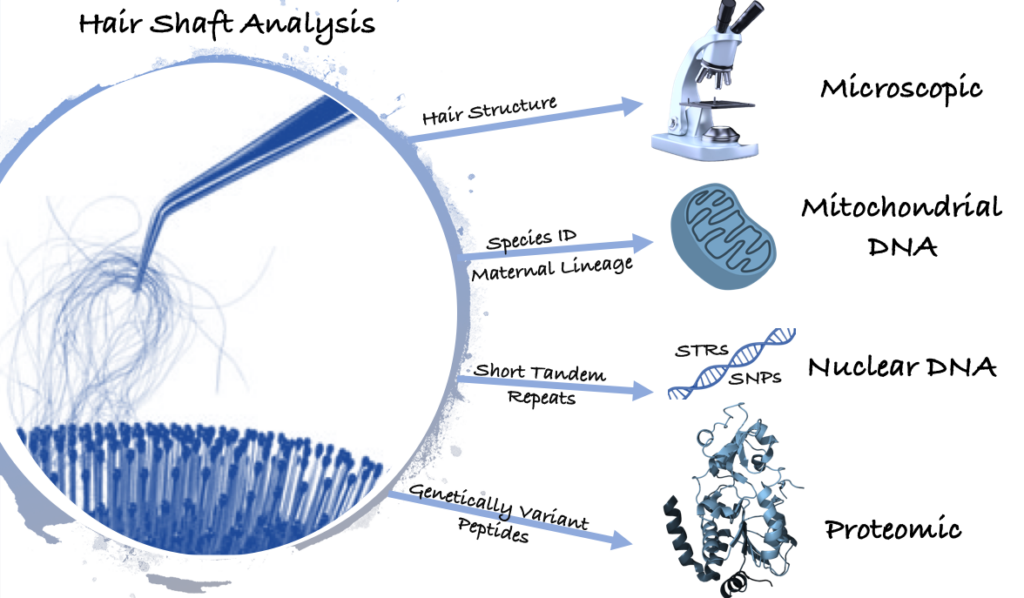
Hair shafts, particularly rootless ones (telogen hair), are common exhibits found in crime scenes. They can be deposited either normally due to shedding or by force due to hair plucking. Based on the American Academy of Dermatology, the normal rate of shed hair per day is 50 to100 strands [1]. However due to the structure and the composition of hair, it was believed for decades that shed telogen hair has no nuclear DNA.
Written by: Mirna Ghemrawi, FLorida International University
Microscopic Analysis
Long before sequencing has emerged as a tool to analyze forensic biological pieces of evidence, the FBI hair examiners used to solely depend on microscopic examination to link a suspect to a crime scene. However, in 2009, the National Academy of Sciences considered that practice as “highly unreliable [2]. By 2015, the FBI concluded that at least 90% of microscopic hair analysis included errors either in testimony or in reports. Because of that, the FBI no longer relies on microscopic analysis but also on mitochondrial DNA testing (mtDNA) [3]. Today, microscopic hair interpretations are rarely used as probative evidence but instead have become an investigative lead.
Mitochondrial DNA Sequencing
Instead, mtDNA analysis can be used to link human hair to a suspect. However, unlike nuclear DNA (nuDNA), mtDNA doesn’t have high discriminatory power, due to its commonality within the maternal lineage. That is, mtDNA is inherited from the mother’s side, with few rare exceptions where the father contributes to the profile. [4]. Hence, the genetic composition of the mitochondria would not differentiate a grandmother from her daughter or her grandchildren, leaving the differentiation aspect to the nuDNA. MtDNA sequencing can also be used to provide substantial information in species identification when examining a miscellaneous hair sample [5]. Since not all hair samples are human in origin, this technique can be used to determine the animal which contributed the hair.
Nuclear DNA Analysis
Nuclear DNA can be found in any nucleated cell in the body from saliva to bone to teeth as well as hair. A small amount of DNA is amplified using a technique known as Polymerase Chain Reaction (PCR), that makes ample copies of few targets known as Short Tandem Repeats (STRs). An STR profile is unique for each individual and is used to search against a database like CODIS to find a match. Hence, being able to get a profile from extracted nuDNA from hair is in fact possible. A study by Opel et al demonstrated partial DNA profiles can be obtained from rootless telogen hair using mini-STR primers [6]. Those primers were placed closer to the repeat region, increasing the potential to amplify STR markers from degraded/fragmented samples (figure 1). In their paper they also collaborated with Eric Buel and Janice Nicklas to develop a degradation index that demonstrated that much of the DNA in hair shafts is highly degraded.

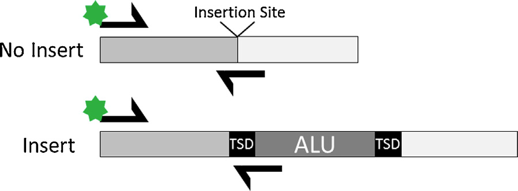
Another study by Grisedale et al shows promising results from rootless hair using an enhanced version of DNA extraction with a novel kit by InnoGenomics named InnoTyper21 (IT21) [7]. This kit has a high tolerance to degradation and inhibition, amplifying short amplicons ranging from 60 to 125bp. It consists of 20 Alu markers that, conceptually, are different from STRs by having two states, insertions or nulls, instead of number of repeats (figure 2). Based on its validation study on five different populations, it exhibits a random match probability of 1 in 3.8 million [8]. Although this is a promising typing method for highly degraded samples, those markers are not in the national DNA database, limiting their usefulness if used solely.
Most recently, a study by Brandhagen et al also demonstrates that nuclear DNA, although fragmented, can be recovered from shed hair [9]. Surprisingly enough, it encompasses the majority of total DNA, further debunking the myth that only mitochondrial DNA can be extracted from rootless hair, and proving that nuDNA is present abundantly when compared to mtDNA. This study, however, used the Whole Shotgun Sequencing method, which is not based on targeted PCR amplification, but rather on shearing the DNA into random fragments and then cloning them into a vector for sequencing. Thus, this method has facilitated a characterization of nuDNA in telogen hairs that wasn’t possible using other techniques, again proving that nuDNA exists in high quantity but low quality. To increase the recovery of nuDNA from hair, they also suggested using extraction protocols that either preferentially recover small fragments or combine more than one protocol to obtain both small and large fragments [9].
In conclusion, these studies totally debunk the myth that there is no nuclear DNA in hair shafts. Instead there is plenty of nuDNA present, but this DNA has become highly degraded during the formation of the hair shaft. Therefore, since nuDNA in shed hair has been characterized to be in high amount but not pristine, more studies should focus on developing assays that particularly tackle these very degraded samples.
Proteomic Analysis
Last but not least, other groups of researchers focus on proteomics, that is the study of proteins to locate genetically variant peptides (GVPs) for human identification. Since DNA degrades relatively faster than proteins, examining the later molecule can complement a partial DNA profile [10]. A group of scientists at NIST have recently published a paper on the identification of GVPs in human hair keratin and discussed the potentials of combining different methods to cover a higher number of variations [11].
References:
[1]https://www.aad.org/hair-shedding
[2] https://www.ncjrs.gov/pdffiles1/nij/grants/228091.pdf
[4] Melton T, Holland C. Routine forensic use of the mitochondrial 12S ribosomal RNA gene for species identification. Journal of forensic sciences. 2007 Nov;52(6):1305-7.
[5] Luo S, Valencia CA, Zhang J, Lee NC, Slone J, Gui B, Wang X, Li Z, Dell S, Brown J, Chen SM. Biparental inheritance of mitochondrial DNA in humans. Proceedings of the National Academy of Sciences. 2018 Dec 18;115(51):13039-44. https://www.pnas.org/content/115/51/13039.short
[6] Opel KL, Fleishaker EL, Nicklas JA, Buel E, McCord BR. Evaluation and quantification of nuclear DNA from human telogen hairs. Journal of forensic sciences. 2008 Jul;53(4):853-7.
[7] Grisedale KS, Murphy GM, Brown H, Wilson MR, Sinha SK. Successful nuclear DNA profiling of rootless hair shafts: a novel approach. International journal of legal medicine. 2018 Jan 1;132(1):107-15.
[8] Brown H, Thompson R, Murphy G, Peters D, La Rue B, King J, Montgomery AH, Carroll M, Baus J, Sinha S, Wendt FR. Development and validation of a novel multiplexed DNA analysis system, InnoTyper® 21. Forensic Science International: Genetics. 2017 Jul 1;29:80-99.
[9] Brandhagen MD, Loreille O, Irwin JA. Fragmented Nuclear DNA Is the Predominant Genetic Material in Human Hair Shafts. Genes. 2018 Dec;9(12):640.
[10] Parker GJ, Leppert T, Anex DS, Hilmer JK, Matsunami N, Baird L, Stevens J, Parsawar K, Durbin-Johnson BP, Rocke DM, Nelson C. Demonstration of protein-based human identification using the hair shaft proteome. PloS one. 2016 Sep 7;11(9):e0160653.
[11] Zhang Z, Burke MC, Wallace WE, Liang Y, Sheetlin SL, Mirokhin YA, Tchekhovskoi DV, Stein SE. Sensitive Method for the Confident Identification of Genetically Variant Peptides in Human Hair Keratin. Journal of forensic sciences. 2019 Oct 31.
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!