Today’s blog is written by guest blogger, Rich Press. Reposted from the NIST blog with permission.
Researchers at the National Institute of Standards and Technology (NIST) have produced synthetic gene fragments from SARS-CoV-2, the virus that causes COVID-19. This material, which is non-infectious and safe to handle, can help manufacturers produce more accurate and reliable diagnostic tests for the disease.
Tests for an active infection — as opposed to antibody tests that indicate a past infection — work by detecting the virus’s genes on a nasal swab. But a negative result does not necessarily mean that a person is disease-free. It could be that the amount of virus is too low for the test to detect, which is especially possible during the first days after catching the virus.
“Having better data on test sensitivity will help us understand how often tests for COVID-19 produce a negative result for people that are actually infected,” said NIST research scientist Megan Cleveland.
To help with this, Cleveland and her colleagues at NIST have produced synthetic fragments of the virus’s genes, which are written in RNA, a molecule that encodes information much like DNA. Synthesizing RNA is not new or groundbreaking. What makes this material notable is that NIST scientists have measured very carefully how many fragments are in each vial they ship.
Using this material, researchers can measure sensitivity by running tests against known quantities of viral RNA. They can also use it to develop more sensitive tests or new types of tests that are faster or easier to administer.
This article describes the synthetic RNA material from NIST and explains how it can be used to measure sensitivity. To understand that, it helps to know a bit about the coronavirus and about how COVID testing works.
PART I: HOW DOES COVID TESTING WORK?
Tests for an active infection work by detecting the RNA of the virus that causes the disease.
RNA is similar to DNA in that both encode information in chemical units that are represented using a four-letter alphabet. DNA’s chemical units are abbreviated as A, C, G and T, while RNA contains the units A, C, G and U.
Viruses can have RNA or DNA-based genes. SARS-CoV-2, the virus that causes COVID-19, is an RNA virus. Here’s an illustration:
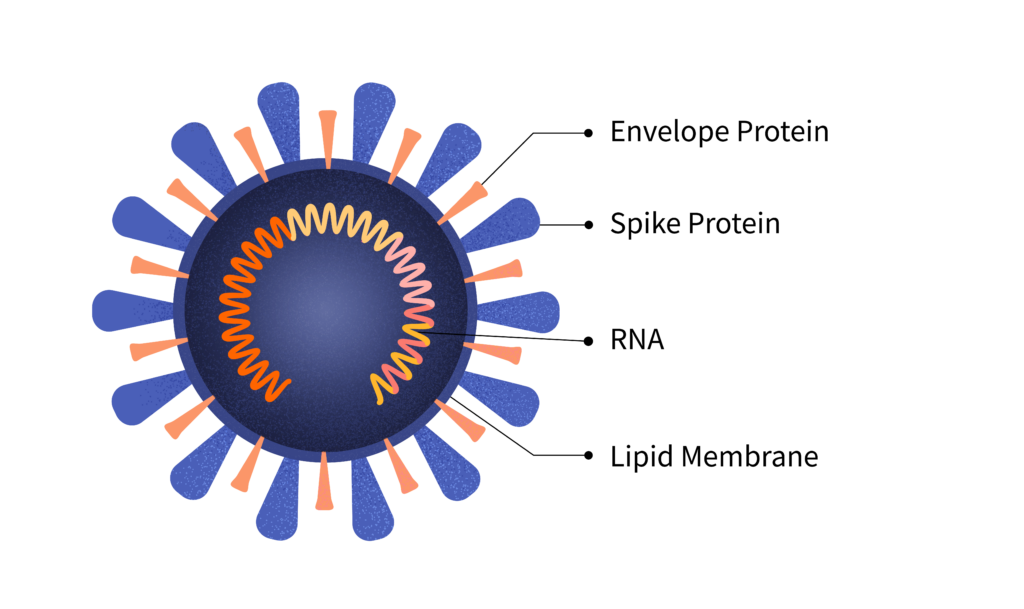
The virus is a tiny, spiky sphere. The RNA, which contains instructions for making more copies of the virus, lies coiled inside it.
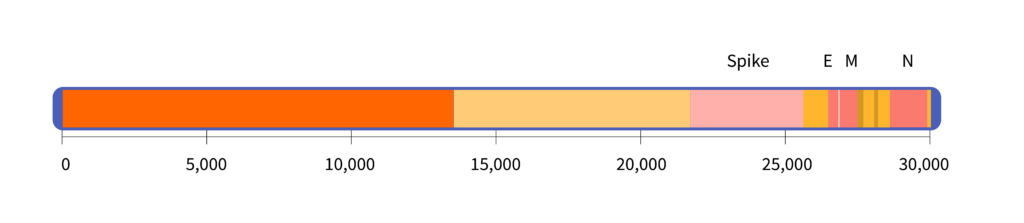
If you were to uncoil the virus’s RNA and spread it out in a straight line, it would look like this:
The virus’s genome is about 30,000 letters long. For comparison, the human genome is roughly 3 billion letters, or 100,000 times longer.
The diagram above shows the location of the gene that codes for the spike protein. It also shows the E gene, which codes for the envelope protein; the M gene, which codes for a membrane protein; and the N gene, which codes for a structure called the nucleocapsid.
Tests for a COVID infection detect specific sequences within the virus’s RNA. For example, one of the tests designed by the Centers for Disease Control and Prevention (CDC) targets a sequence of 72 letters that sits within the N gene:
At 72 letters, this sequence is shorter than a tweet. Yet the only known place where this sequence naturally occurs is in the SARS-CoV-2 virus. If the test detects that sequence, it has detected SARS-CoV-2.
Different tests target different RNA sequences, but they all involve the same steps.
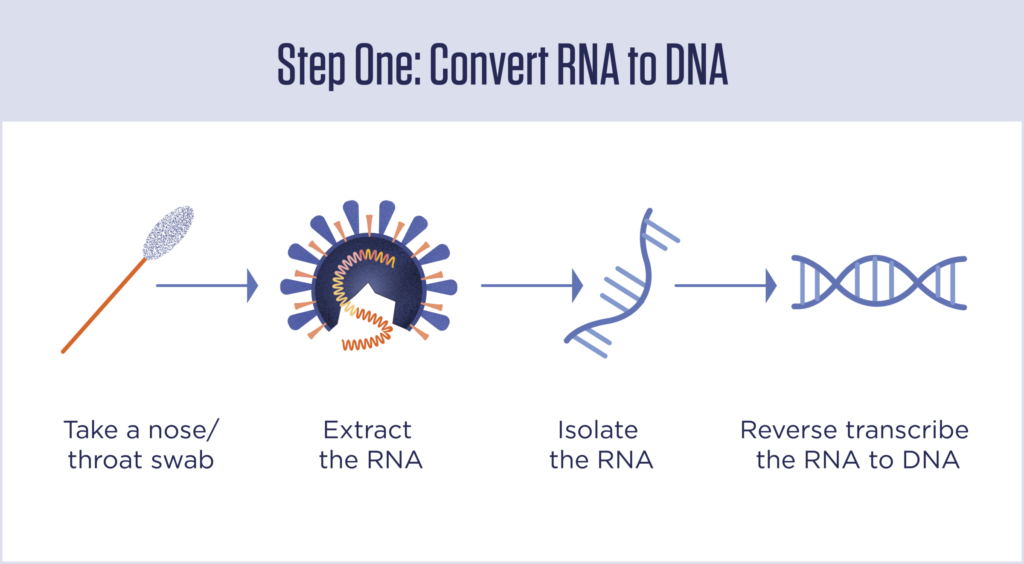
Step 1: Isolate the RNA and convert it to DNA
Most tests begin by inserting a swab into a person’s nasal cavity, where it picks up a mix of human cells, bacterial cells, viruses and whatever else is in there. RNA is then extracted from the sample, and everything else is washed away. Then, short genetic sequences, called primers, are added that will bind with the target sequence if it is present. If the primers find their targets, they kick off a process that transcribes the RNA sequence into a new molecule of DNA. If the primers don’t find their target — that is, if the virus is not present in the sample — nothing happens.
This is called “reverse transcription” because in most cells in your body and in other living things, DNA is transcribed into RNA, not the other way around.
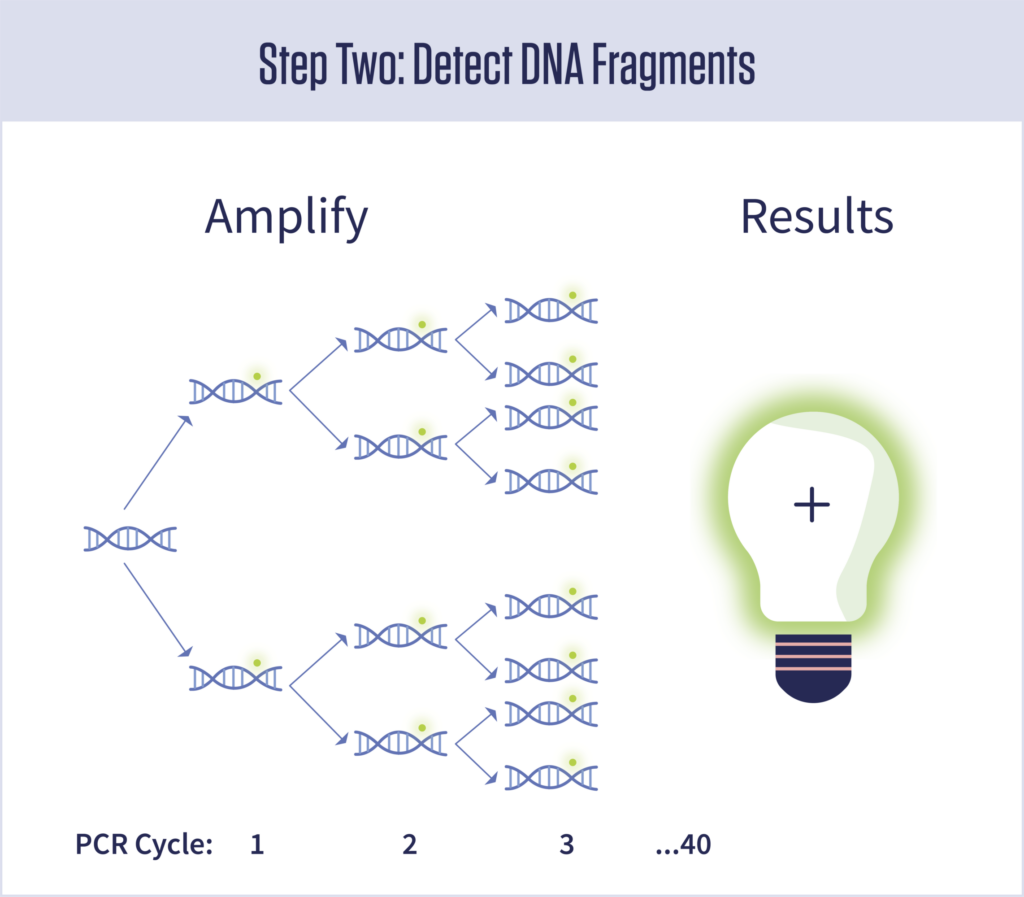
Step 2: Detect DNA sequences
At this point, the sample will contain DNA only if RNA from the coronavirus was present on the nasal swab. However, the amount of DNA will be far too little to detect. The next step is to increase, or “amplify,” the DNA, so it can be detected.
These tests are often called “PCR tests” because they rely on a technique called polymerase chain reaction, which repeatedly doubles the amount of DNA in the sample by copying it. It doubles the DNA, then doubles it again, and so on in a series of chained reactions. Most PCR tests run up to 40 cycles. If you start with only 100 copies of the target sequence (from 100 individual viruses), after 40 cycles of doubling you will have more than 100 trillion copies of it.
PCR is also kicked off by primers and related molecules called “probes” that will only work if they find their targets. If the full target sequence is present, the DNA will be amplified. If not, nothing happens.
The viral tests use a special type of PCR called quantitative, or qPCR, in which a molecule that produces a tiny glow is turned on every time a DNA strand is copied. If in the end you have 100 trillion DNA strands, you also have 100 trillion fluorescent molecules floating in the test solution. If the instrument detects their greenish glow, the test is positive. No detectable glow, it’s negative.
PART II: THE SYNTHETIC VIRAL RNA FROM NIST
The material produced by NIST will allow manufacturers to measure the sensitivity of their tests. In other words, it will help them answer the question: What is the least amount of virus that can be detected by a COVID PCR test?
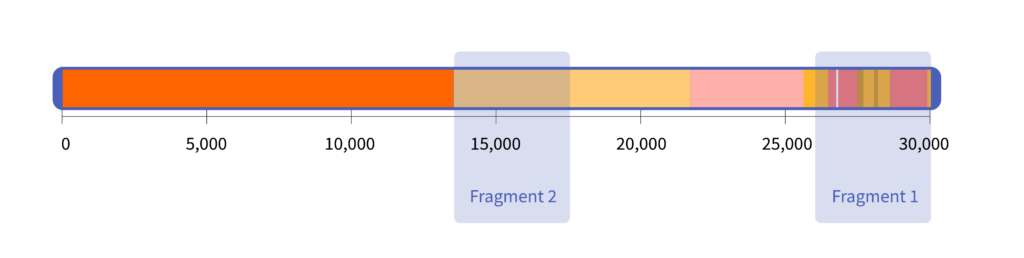
The NIST material contains the two RNA fragments shown here:
Each of these two fragments is roughly 4,000 letters long. NIST researchers chose to synthesize these particular fragments because they contain the RNA sequences targeted by a large number of diagnostic tests, including those designed by the CDC.
These RNA fragments are distributed by NIST in small vials packed in dry ice, along with a data sheet that lists the concentration of the fragments in the solution. The solution contains roughly 1 million copies per microliter, or one millionth of a liter (one drop of water contains about 20 microliters). NIST measured this concentration multiple times using a technique called digital PCR, or dPCR, which counts the number of individual DNA fragments in a volume of liquid.
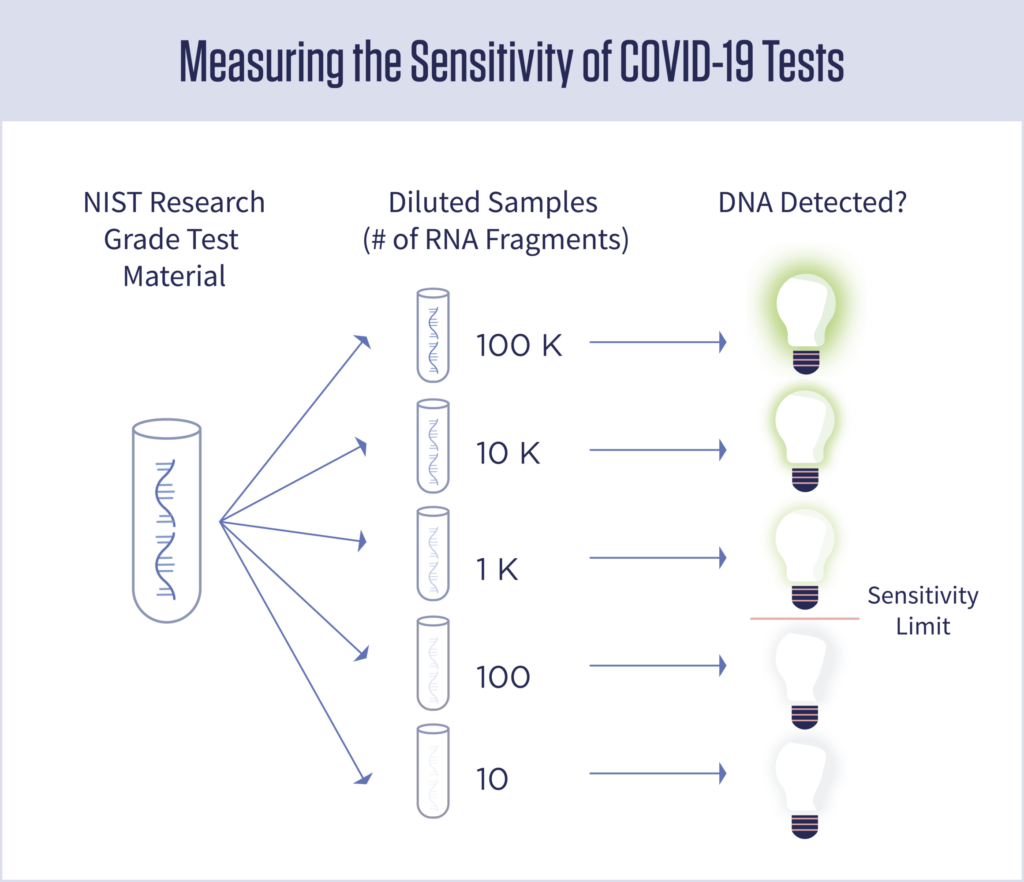
Researchers can use the RNA fragments from NIST to measure the sensitivity of coronavirus tests. To do that, they would use the concentrated solution from NIST to create a series of increasingly diluted samples. They would then run each of those dilutions through their test to find the lowest concentration of RNA fragments that still produces a positive result. Here’s an example:
In this example, the test has been shown to be sensitive enough to detect as few as 1,000 copies of the virus.
In addition to measuring sensitivity, researchers might also use the synthetic RNA from NIST to develop more sensitive tests or new kinds of tests. For instance, recent research has shown that it might be possible to track the spread of COVID-19 in a city by testing municipal wastewater. The material from NIST might help researchers ensure that they can reliably detect very low concentrations of virus in millions of gallons of wastewater.
NIST is releasing this synthetic RNA as a “research grade test material,” though NIST scientists are planning to further develop it into the type of standard reference material (SRM) that NIST is known for. NIST is providing this material at no cost to researchers, test manufacturers and testing laboratories. Technical information and instructions for requesting the material are available on the NIST website.
Genetic supply companies are also synthesizing portions of the virus’s RNA, but what sets the NIST material apart is the amount of data that comes with it. Detailed information on how NIST scientists synthesized the fragments and measured their concentration is available in an online Guidance Sheet. Additional information, including raw data and statistical analyses of the concentration measurements, is available on a GitHub site. NIST is providing this data as part of its mission, as the nation’s measurement laboratory, to advance measurement science.
“Measuring sensitivity is an important part of test development,” said Peter Vallone, the NIST research scientist who oversaw development of the new material. “And we want to make sure that people have the materials and information they need to make the most accurate measurements possible.”
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!