Today’s guest blog is written by Mike Cariola, CEO, Bode Technology and Abigail S. Bathrick, Senior Research Scientist, Bode Technology Reposted from the ISHI Report with permission.
What is FIGG?
Forensic Investigative Genetic Genealogy (FIGG) pairs advanced genotyping technologies with traditional genealogical research methods to provide investigative leads for unknown DNA evidence when CODIS searching is unproductive. It gained worldwide attention as an investigative tool after the 2018 arrest of the suspected Golden State Killer, Joseph DeAngelo. Since then, it has been used to resolve hundreds of cold cases and has become a time-efficient method for identifying missing persons and perpetrators of violent crimes. FIGG’s continued success in resolving high-profile cases and sustained media attention have led to a surge in inquiries and requests from investigators interested in applying FIGG to their cold cases. However, the forensic community has identified the need for further development, assessment, and evaluation of FIGG testing procedures to establish best practices for crime laboratories.

Our Current Research:
Comparative Evaluation of Genotyping Technologies for FIGG in Sexual Assault Casework
Development of best practices must start with a systematic evaluation of the technologies currently used to generate high-density single nucleotide polymorphism (SNP) genotypes. These technologies were developed and validated with pristine, high-yield samples intended for use in clinical genomics, whereas forensic samples typically contain old, degraded, biologically contaminated, and low-template DNA. Our work evaluates how degradation and low-template DNA affect the quality, accuracy, and reproducibility of SNP genotypes and how this affects downstream genealogical database searching and identification of potential relatives.
Three genotyping technologies were investigated in this study: SNP microarray testing with Illumina’s Global Screening Array v2 BeadChip (array), whole genome sequencing (WGS) on the NovaSeq 6000, and targeted sequencing with Qiagen’s ForenSeq Kintelligence Kit on the MiSeq FGx. The technologies were compared for their sensitivity to low-level DNA input concentrations and specificity for degraded DNA. All three technologies were found to be sensitive to low-level DNA inputs. DNA inputs of at least 500 pg generated call rates >85% with both array and WGS analyses, while Kintelligence produced call rates >90% with as little as 100 pg DNA. Comparisons between the technologies demonstrated >95% concordance. Prior to genotyping, DNA degradation was estimated with Quantifiler Trio’s Degradation Index (DI), with DI 1–10 indicating slight to moderate degradation and DI >10 indicating significant degradation. For samples degraded via UV irradiation, increased degradation adversely affected call rates for both array and WGS processing, whereas Kintelligence call rates were robust against degradation (Table 1). While genotype concordance was negatively impacted for array, WGS and Kintelligence demonstrated concordance of >98% and >96%, respectively, regardless of DI value (Table 1).
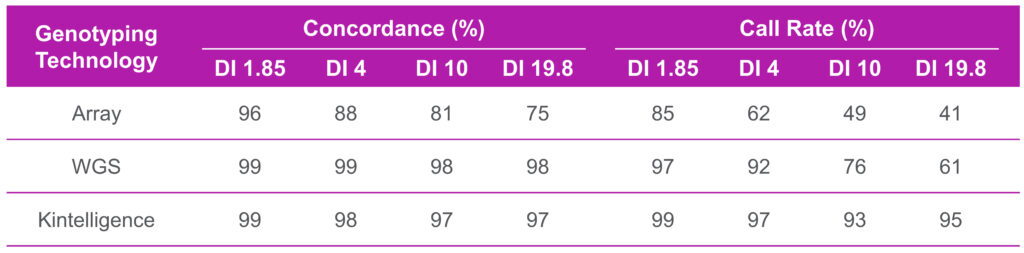
Table 1
The high-density SNP genotypes resulting from the sensitivity and specificity samples were compared against genotypes databased in GEDmatch to determine the maximum distance at which known or potential genealogical associations could be identified. Robust matching was observed out to the 2nd cousin level with low-level DNA inputs for all technologies. Slight to significant degradation (i.e., DI 1.85–19.8) had minimal impact to 2nd cousin matching for Kintelligence and WGS, while moderate degradation (i.e., DI >4) significantly impacted 2nd cousin matching from array processed samples. Additional limitations became apparent during GEDmatch searching/matching. Anomalous results were observed with the WGS-based SNP set of >2 million loci or donors of non-European ancestry, and inconsistencies were observed in genotypes generated with Kintelligence when compared to array and WGS. Kintelligence also displayed limited utility due to its incompatibility with third-party comparison tools.
The limitations of each technology will be further tested by examining challenging samples exhibiting both low-level DNA concentration and degradation using a known donor for whom family members of known relationship distance greater than 5th degree (first cousin once removed) are present in GEDmatch. After genotyping with methods most applicable to each sample’s particular characteristics, a full genealogical investigative workflow will be applied to demonstrate whether increasingly distant relatives can be identified and at what distance identification is no longer possible. Genealogical workflows for the three technologies will also be compared for range and accuracy. All genealogical workflows will conform to the Genealogical Proof Standard, which is a set of criteria used by genealogists to evaluate the quality of their research and the conclusions they draw from it. By adhering to these standards, genealogists can ensure that their research is thorough, accurate, and reliable, leading to more credible genealogical conclusions.
By demonstrating the strengths and limitations of the current technologies, we hope to assist FIGG practitioners in making more informed decisions, optimizing resources, and enhancing investigative outcomes. Additionally, our evaluation may provide support for the development of new genealogical workflows for samples that generate suboptimal results. Such analyses empower forensic experts to select the most effective methods tailored to specific cases, accelerating the identification of suspects and resolution of cases.
This project is supported by Award No. 15PNIJ-21-GG-04143-MUMU, awarded by the National Institute of Justice, Office of Justice Programs, U.S. Department of Justice. The opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect those of the Department of Justice.
Improving FIGG Success with Challenging Forensic Samples
In addition to criminal casework, FIGG has become critical for the identification of missing persons and unidentified human remains. These cases present unique challenges as bone, the primary source of DNA, is highly subject to environmental and chemical damage. Additionally, DNA isolated from bone often contains more exogenous microbial DNA than endogenous human DNA, which can interfere with both microarray analysis and library preparation prior to WGS. In collaboration with the University of Tennessee Knoxville Forensic Anthropology Center, we are using lessons learned from the ancient DNA community to optimize a workflow for library preparation and target enrichment hybridization capture from compromised bone samples that is amenable to implementation in a standard forensic laboratory. With support under NIJ Award No. 15PNIJ-23-GG-04224-RESS, this project aims to improve sequencing of DNA obtained from degraded and contaminated bone samples and ultimately allow for more identifications of human remains.
Our Current Research:
FIGG presents a unique challenge to forensic laboratories, which are not typically equipped with the sequencing technologies used to generate high-density SNP genotypes. Initially, sequencing was outsourced by sending extracted DNA to third-party laboratories specializing in SNP microarray testing and WGS. However, due to the sustained success using FIGG, practitioners are now considering how to integrate end-to-end FIGG processing within an accredited forensic laboratory setting.
Leveraging our years of forensic and sequencing experience, Bode has validated two FIGG sequencing methods to the standards required by ISO 17025: the ForenSeq Kintelligence Kit on the MiSeq FGx and WGS on Element Bioscience’s Element AVITI™ System. The AVITI is a short-read, mid-throughput sequencer that uses Element Bioscience’s novel sequencing-by-binding chemistry to reduce run costs and improve performance when compared to sequencing-by-synthesis chemistry, the current gold standard. AVITI sequencing offers shorter run times and improved base call accuracy and detection, with mean quality scores exceeding Q40 (only one base call in 10,000 is predicted to be incorrect) — a 10X improvement on the current benchmark. Collectively, these innovations have set a new standard for sequencing and make AVITI sequencing ideal for challenging forensic evidence. With this technology, our full-service pipeline now encompasses all steps of the FIGG process: receipt of evidence, sampling, DNA extraction, library preparation, sequencing of SNP genotypes, bioinformatic processing, upload to genealogy databases, genealogical research, and court-ready reporting.
Moving FIGG Forward
Bode is proud to be a leader in the FIGG community. Through ongoing research efforts, we continue to explore FIGG’s potential, contributing new data to the broader body of FIGG knowledge and pushing our work to the forefront of forensic science. We remain committed to advancing FIGG while ensuring the technology adheres to the rigorous standards required for traditional DNA casework methods. Since launching our FIGG service in 2018, Bode has processed over 500 FIGG cases, and 30 cases were processed with our end-to-end FIGG workflow with AVITI sequencing directly following its release. Over 300 investigative leads have resulted from this work, underscoring FIGG’s value to modern forensic investigations. We look forward to the future of FIGG and plan to continue sharing the results of our ongoing FIGG research with the wider community.

