Everyone’s DNA is slightly different. Thirty-five years ago, when scientists invented a way to distill those differences into a DNA profile, they revolutionized forensic science. That profile—a sort of genetic fingerprint—gave investigators a new and extremely reliable tool for solving crimes.
DNA profiling can be so powerful and has been used successfully in so many cases that some people think it is nearly infallible. But the reliability of DNA profiling varies, and other things being equal, it is most reliable when the evidence contains plenty of DNA from just one or two people. That’s why homicides and sexual assaults—crimes that tend to produce a lot of this type of DNA evidence—made up the bulk of DNA casework for many years.
But in the last decade or so, forensic experts have been analyzing DNA mixtures, which occur when the evidence contains a mixture of DNA from several people. They are also analyzing trace amounts of DNA, including the “touch DNA” left behind when someone touches an object. These types of evidence can be far more difficult to interpret reliably than the DNA evidence typical of earlier decades. With old-school DNA, the results tend to be clear cut: either a suspect’s DNA profile is found in the evidence or it isn’t, and nonexperts can readily understand what that means. With DNA mixtures and trace DNA, the results can be ambiguous and difficult to understand, sometimes even for the experts.
These more complex types of evidence make up an increasing share of the DNA casework in the United States, and labs are rapidly adopting new methods and tools to deal with them. More than just an upgrade to the existing tool set, these changes represent a fundamentally new approach to DNA evidence and mark a profound shift in the field. Given the great weight that DNA evidence carries in the courtroom, it is important that lab analysts, criminal investigators, judges, attorneys—and anyone who might sit on a jury someday—understand these changes. So here’s a quick primer on DNA mixtures and trace DNA, what makes them difficult to interpret, and what these changes mean for the future of the field.
Written by: Rich PRess, NIST (REPUBLISHED WITH PERMISSION)
Why have DNA mixtures and trace DNA become so prevalent?
DNA methods have become extremely sensitive. Forensic scientists once needed a relatively large amount of material, such as a visible blood or semen stain, to produce a DNA profile. Today, they can generate a profile from just a few skin cells that someone left behind when touching an object or surface.
This capability is an incredible technological achievement. It is also a positive development because it allows forensic science to help solve a greater variety of crimes. Investigators might be able to solve a sexual assault, for instance, even if very little DNA were recovered. They might investigate a break-in by swabbing the pry bar that was used to force a door, or they might swab a firearm that was used to commit a crime.
But such high sensitivity is a double-edged sword. We often shed small amounts of DNA when we talk, sneeze and touch things. As a result, many surfaces are likely to contain mixtures of minute amounts of DNA from several people. These mixtures have always been present at crime scenes, but when sensitivity was lower, they wouldn’t have been detected or, if they were, labs would not have attempted to interpret them. That is no longer the case.
Are all DNA mixtures difficult to interpret?
Some mixtures are relatively easy to interpret. Others are more complex and require greater care. Still others may be too complex to reliably interpret at all. It depends on the specifics of the case.
Three main factors determine the complexity of a mixture.
- How many people contributed DNA to the mixture? More contributors make a mixture more complex, and therefore, more difficult to interpret.
- How much DNA did each person contribute? Even if a mixture contains plenty of DNA overall, one or several people might have contributed only a tiny amount. The lower those amounts, the more complex the mixture.
- How degraded is the DNA? DNA degrades over time and with exposure to the elements. This can also increase complexity.
When does a DNA mixture become too complex to reliably interpret at all? Currently, there are no established standards for deciding this. Different labs have different protocols. When confronted with a particularly complex DNA mixture, some labs will try to interpret it and others won’t.
Why are complex DNA mixtures difficult to interpret?
To answer this question, it helps to know a bit about DNA profiles. When generating a DNA profile, forensic scientists don’t analyze the entire genetic sequence. Instead, they look at roughly 40 short segments of DNA that vary from person to person. Those different variations are called alleles, and the key to knowing a person’s DNA profile is knowing which alleles they have.
To find out, forensic scientists need enough genetic material to analyze, so they make millions of copies of the alleles. After “amplifying” the DNA in this way, scientists run the alleles through an instrument that sorts them the way a coin counter sorts coins. Instead of ending up in coin slots, the alleles end up as peaks on a chart. The positions of those peaks indicate which alleles are present—that is, they determine a person’s DNA profile.
When the evidence contains plenty of DNA, those peaks are often easy to read. For example, imagine a case in which a killer cuts himself on the knife he used as a weapon, leaving drops of blood at the scene. Analysts have created a DNA profile from those blood drops. In addition, the police have arrested a suspect, collected the suspect’s DNA and generated a profile from it. Here’s what the two profiles might look like:
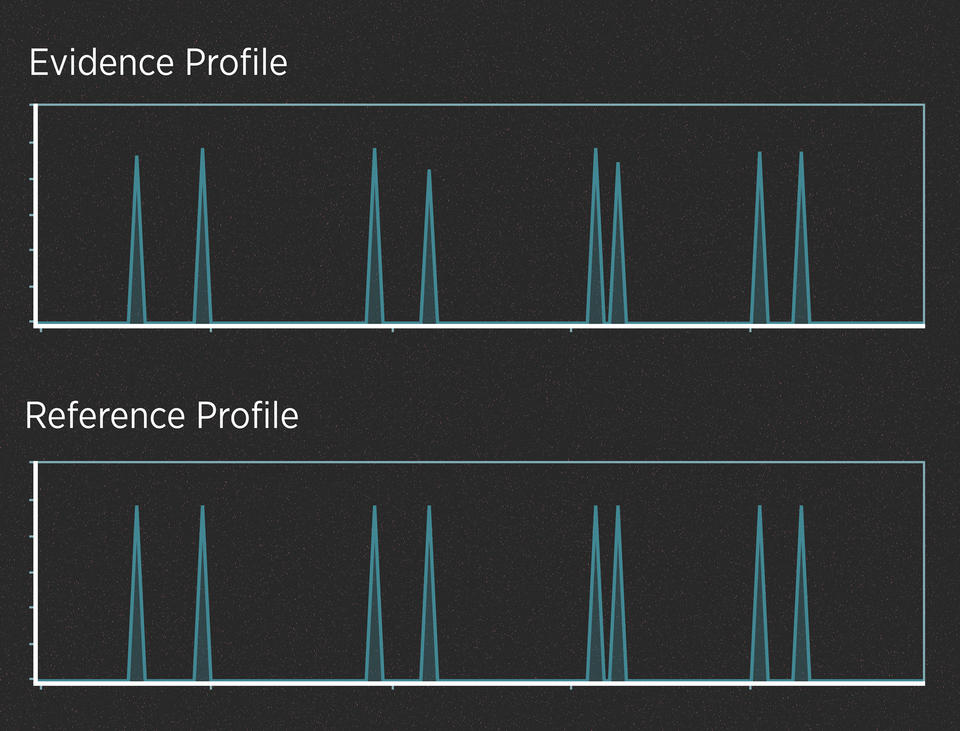
Alleles are said to match when their peaks fall at the same left-to-right position on the chart. When comparing profiles from unrelated people, it wouldn’t be unusual to find that they have a few matching alleles, just as it wouldn’t be unusual to match one or two numbers in a lottery. But it would be incredibly unlikely for all the alleles to match.
So, do the two profiles in the graphic above match? They do, and the DNA analyst would use well-understood statistics to calculate the strength of that match. This analysis does not require much interpretation. In contrast, evidence that contains trace amounts of DNA or a DNA mixture can require a lot of interpretation.
For instance, imagine that the killer in the case above didn’t cut himself and leave drops of blood at the scene. However, investigators recovered the knife and swabbed the handle hoping to find touch DNA. The profiles might look like this:
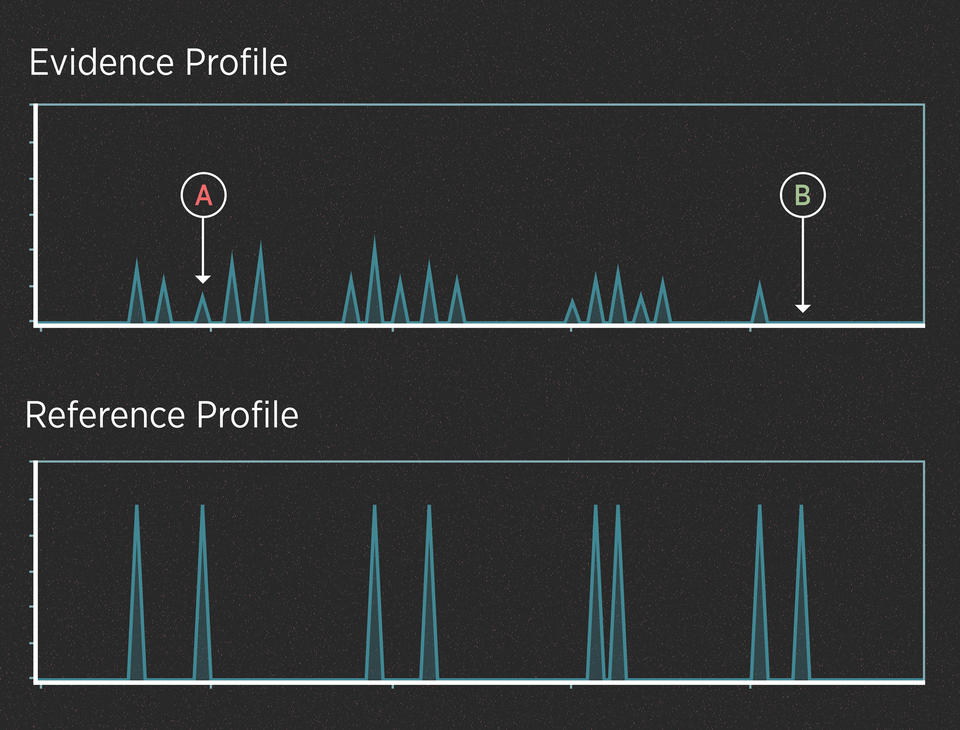
In this case, we can assume from the number of peaks that the evidence contains DNA from more than one person. The peaks are small because the amount of DNA is low. Interpreting this profile involves at least two uncertainties.
UNCERTAINTY #1: When is a peak a peak?
When the amount of DNA is very low, the peaks can be very small. Some peaks can be so small that they disappear entirely (they “drop out” of the profile). Also, small blips in the data can be mistaken for real peaks (they “drop in” to the profile). Many of these effects are random, and they can make it difficult to interpret the evidence.
UNCERTAINTY #2: Whose peak is it anyway?
When analyzing a DNA mixture, the alleles from all the contributors show up on the same chart. This can make it difficult to tease apart the DNA profiles of the individual contributors. To understand why this makes things complicated, recall that after amplifying the DNA, the forensic scientist has a test tube with millions of copies of the alleles in solution. Think of that test tube as a bowl of alphabet soup.
In this bowl of soup, each letter represents a different type of allele. Our suspect is named JOHN Q SUSPECT.

We analyze the soup and find that all the letters in the suspect’s name are present. Does that mean someone named JOHN Q SUSPECT contributed to the soup?
Not necessarily. There could have been two contributors named PATRICK QUEEN and JUSTIN OHR. In that case, the soup would have all the letters needed to spell JOHN Q SUSPECT, even though no person with that name contributed to the soup.

This illustrates an important point about DNA mixtures: Just because a person’s alleles appear in a mixture does not mean that person contributed to it. The alleles may have come from some combination of other people who, between them, have all the allele types in the suspect’s profile.
Because of these uncertainties, it can be difficult to know whether a suspect might have contributed to a mixture. Instead of a simple yes or no, the answer is often expressed in terms of probabilities.
What is probabilistic genotyping software, and how does it help?
Scientists have developed computer programs to help interpret complex mixtures. Probabilistic genotyping software (PGS) uses statistical and biological models to calculate probabilities. For instance, the software is designed to account for drop-in, drop-out and other effects by using mathematics to approximate what happens in a real mixture. PGS also considers the fact that some alleles are more common in the population than others, just as the letter J is more common in peoples’ names than the letter Q.
After computing these probabilities, the software produces a number called a likelihood ratio. That number is the software’s estimate of how much more or less likely it is to see that mixture if the suspect did contribute to it than if the suspect didn’t. The jury may then take that number into account, along with other evidence, when deciding guilt or innocence.
In many cases, mixtures can be interpreted more reliably with PGS than without it, if the analyst understands the assumptions made by the software and the underlying mathematics. This makes PGS an extremely important tool, and one that can help investigators solve many crimes that might otherwise go unsolved.
However, the type of software used, how the software is configured, and which models the software runs can all affect the results. Therefore, different labs might produce different results when interpreting the same evidence. Sometimes those differences can be large enough to call into question the reproducibility of the results. This highlights the fact that every scientific method has its limits, and some mixtures will be too complex to reliably interpret even with PGS. Currently, there is no consensus on how to identify those limits.
Finally, while PGS interprets DNA profiles, it does not address an important uncertainty associated with DNA mixtures and trace DNA.

How confident can one be that the DNA is related to the crime?
While PGS can tell you who might have contributed DNA to a mixture, it can’t tell you how or when their DNA got there. If the evidence contains a lot of DNA, this might not be a problem. For instance, investigators at the scene of a home invasion and homicide might find a broken window with blood on the glass. In that case, they might reasonably conclude that the killer broke the window to enter and cut himself on the way in. In other words, they can associate the DNA in the blood with the crime.
However, if the killer entered through an unlocked door, a swab of the doorknob might yield DNA from many innocent people who, in touching the doorknob, transferred their DNA to it. In addition, DNA can be transferred multiple times. For example, if you shake the hand of a person who later touches the door knob, your DNA can end up on the door knob even though you never touched it. Scientists call this “secondary transfer.” Situations like these show how it can sometimes be difficult to know if trace amounts of DNA are related to the crime.
Scientists have conducted studies to better understand the factors that make DNA transfer more or less likely. They have found that some people tend to shed more DNA than others, and some objects and materials are particularly good vehicles for transferring DNA. Still, our understanding of how, and how often, DNA transfer happens is limited.
When using high-sensitivity methods, however, forensic scientists are more likely to detect and get profiles from irrelevant DNA. That means that the risk of incorrectly associating a person with a crime has gone up in recent years. Sheila Willis, a guest researcher at NIST and the former Director General of Forensic Science Ireland, says that mitigating that risk is especially important when dealing with samples containing very small amounts of DNA. One way to do that, she says, is to consider the totality of the evidence in a case rather than relying solely on an isolated fragment of DNA that might not be relevant.
Should labs just stop analyzing complex DNA mixtures altogether?
No. These types of samples, though often challenging, can still provide very powerful and reliable evidence. If there’s one thing you take away from this explainer, it should be this: methods for interpreting DNA mixtures are not inherently reliable or unreliable. Mixtures exist on a spectrum, and the ability to reliably interpret a particular mixture depends on the specifics of the case.
The key is to ask the right questions. How complex is the mixture in terms of number of contributors and the amount of DNA from each? How confident can we be that the DNA is relevant to the case? What other types of evidence exist to corroborate the DNA evidence? Perhaps more than at any time since forensic DNA methods were invented 35 years ago, this type of critical thinking is needed.
It’s also important to understand the limits of scientific methods. How far can we push new methods when interpreting complex DNA mixtures? How can we establish consistent protocols for deciding when a mixture is too complex to interpret reliably? What additional training do forensic analysts need to use new methods appropriately? NIST is conducting a study that evaluates these issues. Called DNA Mixture Interpretation: A Scientific Foundation Review, this study evaluates the science behind these methods and identifies areas for future research. If you’d like to receive a notice when the study is published later this year, sign up for our email list.