Today’s guest blog is written by Sanne Aalbers, University of Washington Department of Biostatistics
Forensic genetics is concerned with the matching of genetic profiles from evidence and from persons of interest. Match probabilities are usually obtained on a single-locus basis and combined over markers by multiplying over loci. However, this approach assumes that the loci are independent. If this assumption is violated match probabilities may be underestimated, potentially leading to overstating the weight of the evidence. As part of my dissertation, I have examined this property for sequence-based profiles.
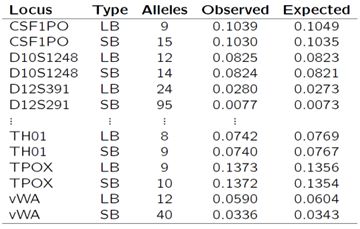
Our data consist of 1,386 samples sequenced over 27 autosomal loci using Illumina’s MiSeq FGx and ForenSeq DNA Signature Prep Kit. I calculated expected matching proportions on a single locus basis using joint genotypic probabilities, assuming Hardy-Weinberg equilibrium (HWE), and allele frequencies as estimated from the data. I then multiplied these with the number of pairs of individuals and compared the results to the observed number of matches. As expected, matching proportions are smaller for sequence-based (SB) data as compared to length-based (LB) data. The decrease is more dramatic for loci that show more sequence variation (Table 1).
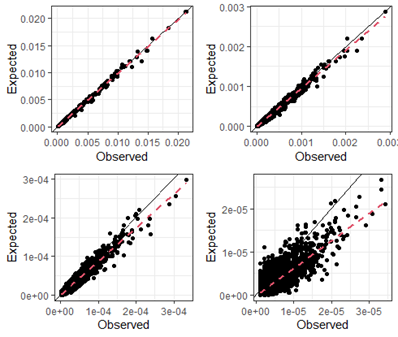
The power of DNA profiling comes from using many loci. To predict multi-locus match probabilities, it is common to use the product rule. The product rule combines single-locus match probabilities by multiplying over markers, assuming that the loci are independent. However, autosomal match probabilities for STR profiles violate the assumption of independence, which increases with the number of loci. Figure 1 displays results for sequence-based data and shows that the product rule may underestimate match probabilities, potentially leading to overstating the evidence. Results for 2 and 3-locus matches (top) show good agreement between the observations and our expectations. However, when going to 4 and 5-locus matches (bottom) results become less and less conservative.
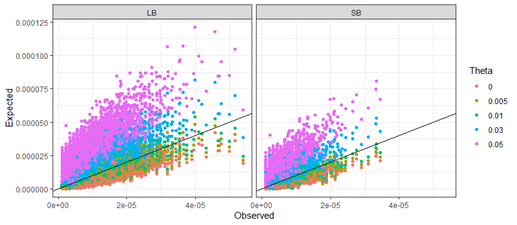
Interestingly, the problem seems to be exacerbated for sequence-based data as compared to length-based data. For 5-locus matches the proportion of conservative results is 28.6% for length-based data, while this proportion decreases to 12.3% for sequence-based data (Table 2). Luckily, there is a solution to this problem. We can compensate for multi-locus dependencies using the theta-correction as outlined in Recommendation 4.10 from the 1996 NRC report. It is common in forensic DNA evidence evaluations to use a theta value around 0.03. Based on 5-locus matches, the proportion of conservative results for length-based data increases from 28.6% using no theta-correction, to 87.5% when using a theta value of 0.03. For sequence-based data, the proportion increases from 12.3% to 64.7%. Using a theta value of 0.05 increases the proportions for both length-based and sequence-based data to over 90% (Figure 2 and Table 2).
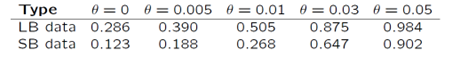
It is important to remember that for forensic evidence evaluations involving autosomal STR markers, the more loci we include the more we underestimate true matching proportions when using the product rule. Our work shows that results are less conservative for sequence data as compared to length-based data. We can invoke a theta-correction to compensate for these multi-locus dependencies. Overall, our work impacts the forensic science community by improving our understanding of the effect of sequence data on match probabilities, a measure integral to DNA evidence evaluations.
I also presented this work as a poster during the ISHI conference and am happy to share it with those interested in more information. I will continue this work in the next months and hope to publish the results as part of my dissertation work. This project was funded by grant 2020-R2-CX-0040 from the U.S. National Institute of Justice. Our work was in part supported by the U.S. National Institute of Standards and Technology.
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!