Today’s guest blog is written by Charissa van Kooten, Netherlands Forensic Institute
Hi there, my name is Charissa van Kooten. My journey in forensic science started over 15 years ago when I followed the inaugural master forensic Science at the University of Amsterdam. Fresh from university I was lucky to be employed by the Netherlands Forensic Institute and went into training to become a reporting officer on biological traces, DNA and DNA kinship. During the last 15 years I saw grow and development in my area of expertise in many ways. One of them was the digitization. From a case file on paper and manual comparisons of SGM+ profiles to digital case files and DNAxs[1] (our data management software system including probabilistic genotyping). These developments had a lot of benefits; better ways to keep the quality high, less subjective decision making, better statistical basis, higher throughput to name a few. It also brought new opportunities because data is now stored digitally.
At the end of 2020 my colleague Corina Benschop from our Research and Development group asked the reporting officers for input concerning a new project that was about to start called: Next Generation DNA decision making. The idea was to explore if it was possible to use data from previous cases to gain insight in choices and their outcomes to help making decisions in future cases. Actually upscaling what we did as reporting officers. Using your experience and that of your colleagues to get the best result. I was interested from the beginning. This mainly had two reasons. One, why wouldn’t you use (more) date. And two, Covid. Which reporting officer doesn’t know the feeling when you look at a low template mixture. What to do? Will it get better after rework? What type of rework? Because of the pandemic we worked partly from home and although you could call a colleague by phone or Microsoft teams for quick advice it is easier when you sit next to each other or in the same hallway. So I volunteered to be a member of the project team.
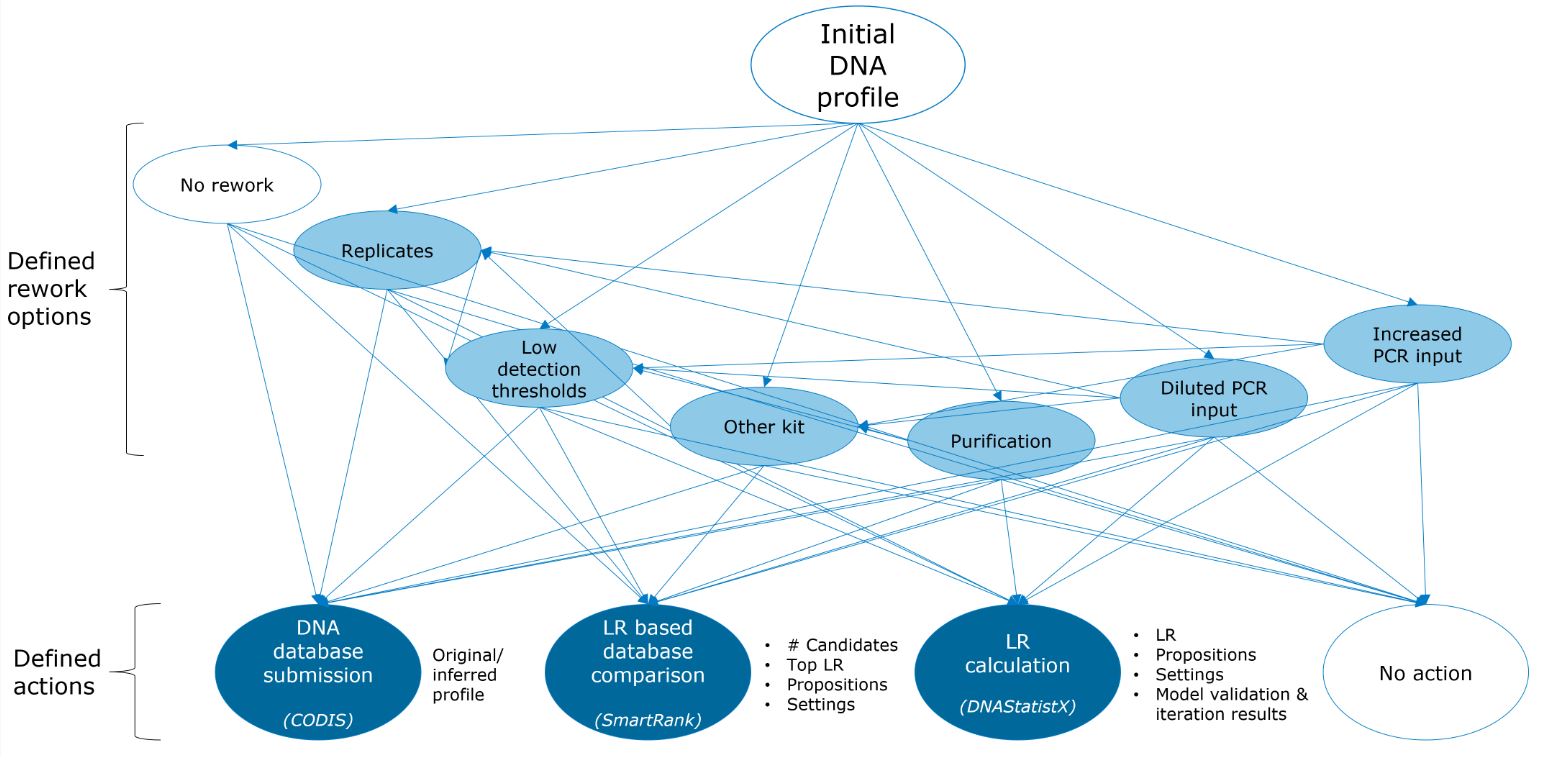
After receiving finances from the Innovatie platform SRK in The Netherlands, we had three agile sprints in 2021 and two in 2022. In these sprints of each three weeks, a project team of data scientists, reporting officers and research scientists developed the DNA decision support tool. Pseudonymized data on DNA profiles and cases were collected from our LIMS system and DNAxs, spanning cases from April 2019 to September 2022. Trace DNA profiles were categorized as being the initial generated profile for a sample (n = 41,392), or resulting from any type of rework (n = 9,906 profiles). Furthermore, we gathered information on three different actions that may be performed with a DNA profile, with or without the use of rework: 1) DNA database submission (CODIS), 2) DNA database search (SmartRank[2]), 3) LR calculation (DNAStatistX[1]).
Fig. 1 provides an overview of the data collected in the DNA decision support database. To enable searching for similar DNA profiles using this tool, various parameters were computed. These were: Pre-profile parameters (such as PCR input in µL); Allele count parameters (such as maximum and total allele count); Peak height parameters (e.g. whether avg. height is higher or lower than the stochastic threshold); Drop-out and degradation parameters (e.g. degradation slope, number of locus drop-outs); LoCIM parameters (e.g. number of inferred alleles); Male count parameters (e.g. X/Y imbalance). The Search dashboard of the DNA decision support tool allows uploading a DNA profile of interest and its corresponding parameter values are computed and presented.
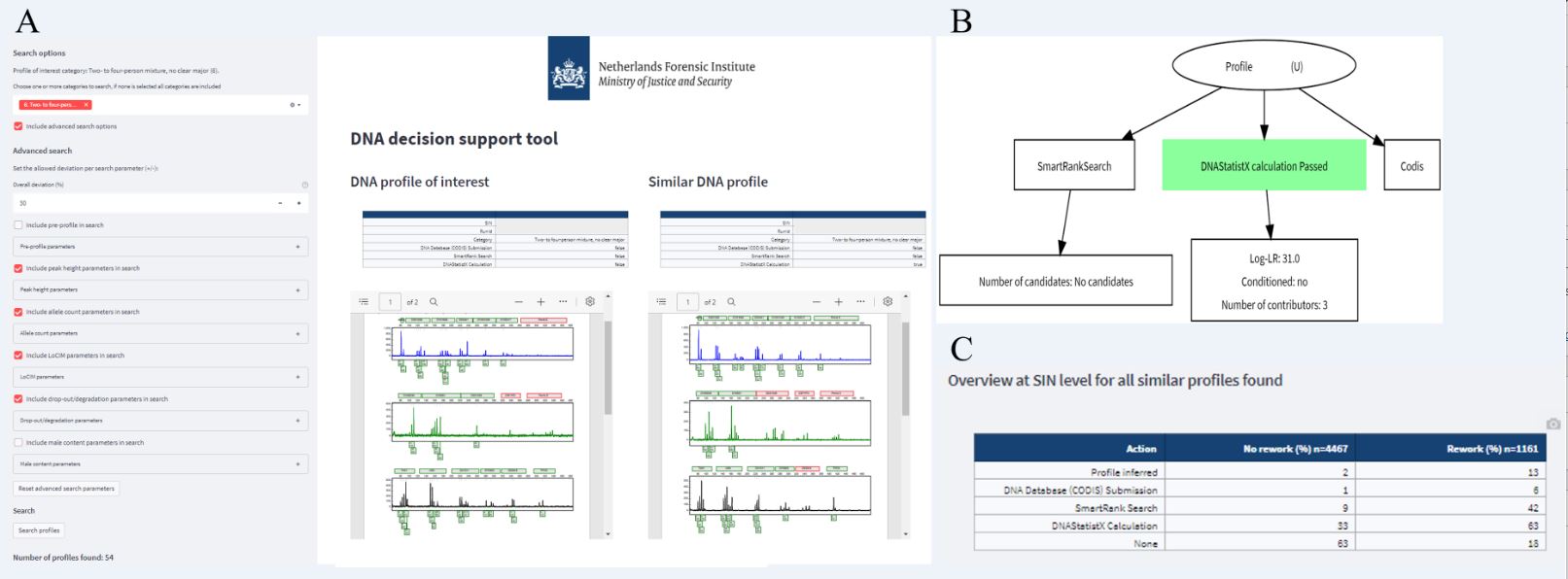
Insight in actions with similar profiles is then provided through an activity graph per similar profile and an overview table with percentages of similar profiles that were used for a specific action (examples in Fig. 2). So far the DNA decision support tool provides insight in the decision process in casework, leveraging thousands of previous casework profiles, actions with these profiles and their outcome. This work on the DNA decision support tool was summarized in the Poster I presented at the ISHI 33 called ‘Using previous DNA casework data to aid decision making in the process of DNA profile interpretation’ and was also presented in [3].
So what’s next. The coming year we hope to develop a live connection between our DNA decision support tool and our casework systems, to add pre-profile information about the purpose of the DNA investigation, to gain more insight in prediction success rates for any of the three actions given the information in the case and to train our reporting officers in the use(fulness) of the tool.
Last but not least, this project would not have been possible without the skills, knowledge and enthusiasm of all project members, and I am grateful to be part of the team.
[1] C.C.G. Benschop, J. Hoogenboom, P. Hovers, M. Slagter, D. Kruise, R. Parag, et al., DNAxs/DNAStatistX: Development and validation of a software suite for the data management and probabilistic interpretation of DNA profiles, Forensic Sci. Int. Genet. 42 (2019) 81–89.
[2] C.C.G. Benschop, L. van de Merwe, J. de Jong, V. Vanvooren, M. Kempenaers, C. van der Beek, F. Barnie, E. L´opez Reyes, L. Moulin, L. Pene, P. Gill, H. Haned, T. Sijen, SmartRank: a likelihood ratio software for searching national DNA databases with complex DNA profiles, Forensic Sci. Int. Genet. 29 (2017) 145–153.
[3] C.C.G. Benschop, C. van Kooten, D. Zandstra, P. Sjoukema, F.E. Duijs, M. van den Berge, C. Schepers, J. Mandersloot, R. Ypma. Using previous DNA casework data to aid decision making in the process of DNA profile interpretation, Forensic Sci. Int. Genet. Suppl. Ser. (2022) In press. DOI:10.1016/j.fsigss.2022.10.052
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!



