Today’s blog is written by Curt Hewitt, PhD, Signature Science, LLC. Read the full version in the November 2020 issue of The ISHI Report.
Introduction
A crime scene. A suspect. A brass shell casing to tie them together. The trace amount of DNA collected is too degraded for DNA profile generation. With that, the suspect walks away, free to strike again. Reaching a dead end like this one in the forensic laboratory is not uncommon and frustrates even the most seasoned analysts and law enforcement officials. In recent years, however, the use of next generation sequencing and genetic genealogy techniques to generate investigational leads have shown us that advancements in forensic science can inject hope into cold cases. In the case of our shell casing, can we find another way in the forensic laboratory to exploit the sample and identify the suspect? Protein molecules, abundant and relatively robust against degradation in touch samples offer significant potential. Signature Science’s team has been working to develop methods to enable protein sequencing for human identity analysis with funding from the U.S. government’s Intelligence Advanced Research Projects Activity (IARPA’s) Proteos program. Based on our research to date, protein sequencing has significant potential as a forensic tool.
Background
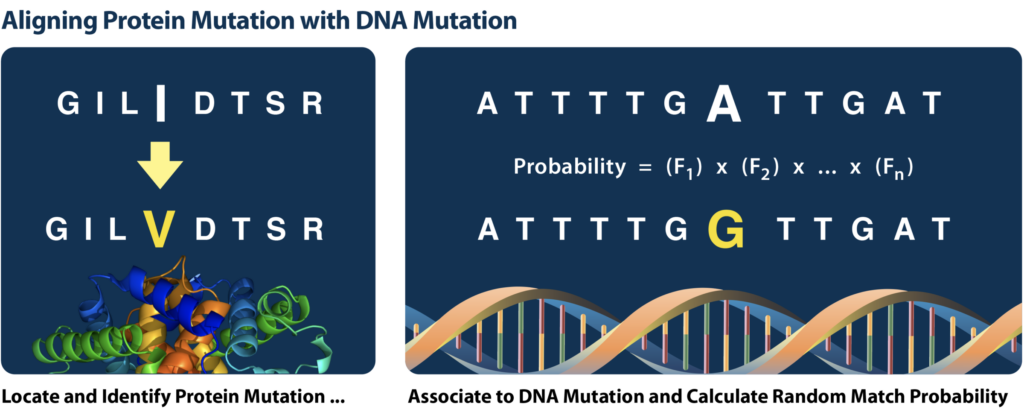
Pioneers of protein sequencing for human forensics, or forensic proteogenomics, showed that polymorphisms in the amino acid sequence of proteins could be used to infer the underlying single nucleotide polymorphisms (SNPs) which gave rise to them during the process of transcription and translation.1 In essence, these protein variants can be used as a SNP genotyping tool complementary to other forensic SNP genotyping methods like DNA sequencing or microarrays. Early protein sequence characterization focused on rootless hair shafts, a forensic sample type notorious for poor DNA yields. More recently, bone, teeth, fingernails, and even archeological samples containing badly degraded DNA have been studied.
Touch Samples
Signature Science’s IARPA-funded work focuses on touch samples which, while occasionally containing sufficient DNA to produce a complete STR profile, more often result in no profile or in a complex mixture of partial profiles. The difference between touch samples and other common forensic samples, such a blood, cannot be overstated, as significant portion of the DNA in touch samples is extracellular.2 Unlike DNA present in samples like dried blood, which remain encapsulated in lipids and protein, much of the touch DNA we collect has been exposed to harsher environments which include nucleases, UV radiation, and reactive chemical species. A significant number of the cells left behind on a touch sample, conversely, lack DNA. These keratinocyotes lose their nuclei as they transit to the skin surface, resulting in a dead, dried cell composed primarily of protein and lipids.
Sample Collection and Extraction
Developing an extraction protocol to enable both DNA and protein analyses presented challenges. Signature Science’s team faced two largely incompatible workflows to capture two different biomarkers from the same sample. Forensic DNA extraction methods focus on maximizing DNA yield. To free the DNA from other cellular material, strong surfactants and enzymes are employed to degrade nucleases and structural proteins. Protein sequence analysis is impossible following this step. Conversely, if a laboratory were to prioritize protein analysis, the harsh denaturation conditions would likely degrade any DNA present in the process. A simple approach to enable parallel DNA and protein analysis would be to split the sample in half; however, a 50% decrease in DNA yield from a trace sample can be the difference between success and failure in generating a DNA profile. For this reason, it is not feasible to expect forensic laboratories to sacrifice potential DNA yield based on the chance that a set of protein markers may also be collected.
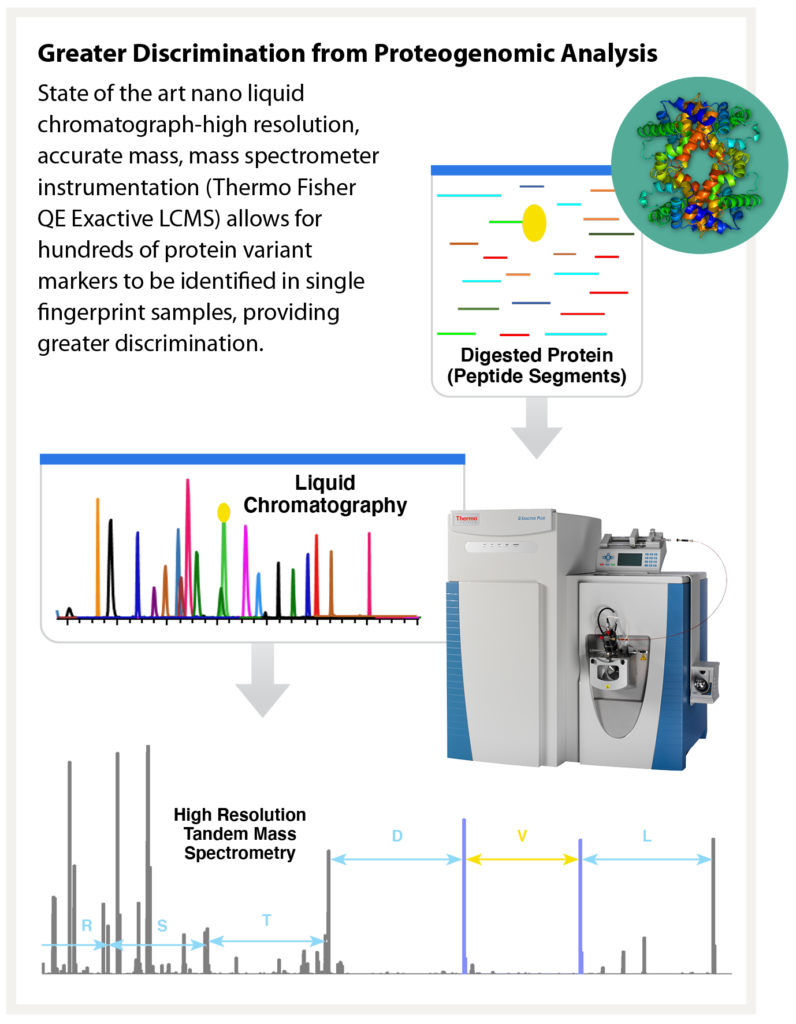
Signature Science’s team worked for more than two years to develop collection and extraction protocols which collect DNA with equivalent quality, quantity, and methodologies currently used in forensic laboratories.3,4 The result is a workflow that enables protein to be recovered from fractions of the sample previously discarded as waste. Better still, this recovered protein can be stored frozen while traditional DNA analysis takes place. If the resulting DNA profile is sufficient for matching, the protein can be discarded. If insufficient DNA is collected, the lab has the option to thaw and analyze the protein. We routinely produce robust protein marker profiles (more than 60 markers per sample) from as little as 1 microgram of protein collected from a touch sample.
Sample Analysis
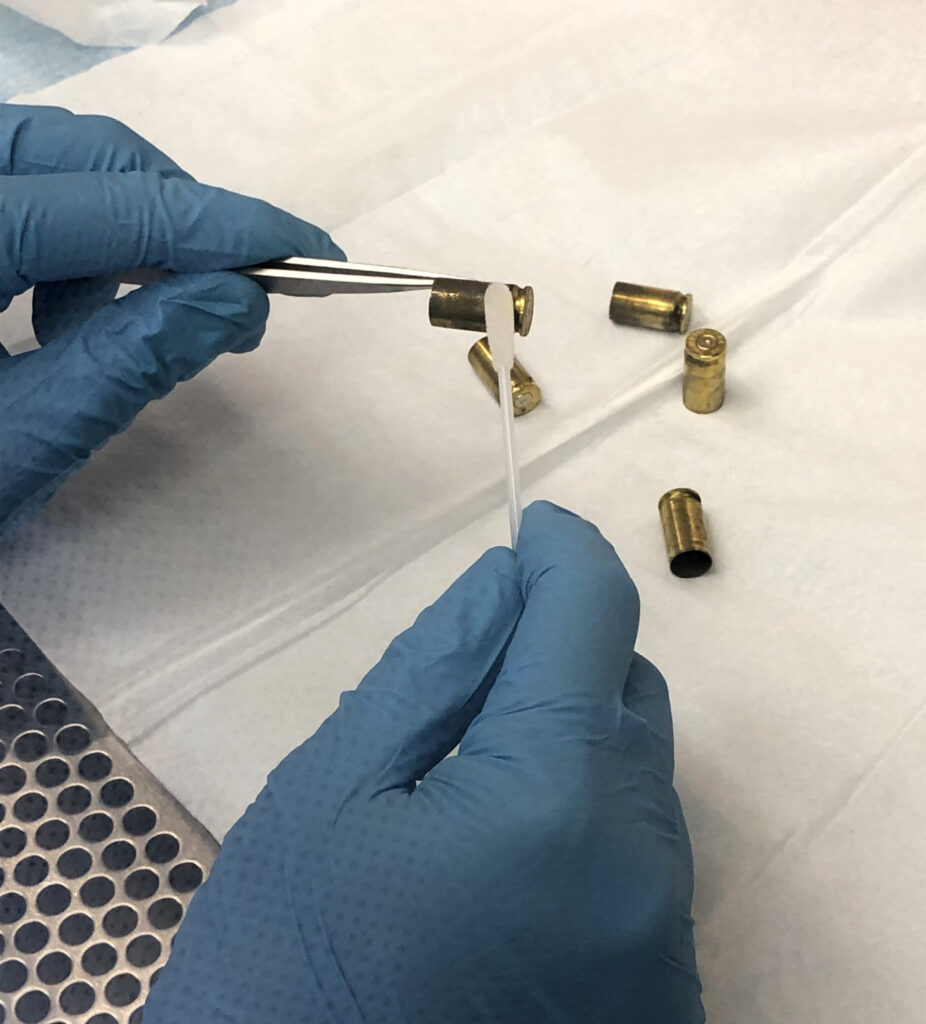
As part of the Proteos program, Signature Science has analyzed numerous touch samples prepared both internally and by an independent third party that were deposited across a range of nonporous substrates such as glass and plastic, porous surfaces like laminate or wood, or even some especially challenging forensic sample types like brass shell casings. Our proteomic analysis methods have routinely resulted in the accurate identification of the correct contributor from a panel of 51 potential donors from as little as 140 ng of total protein, a common yield for even trace touch samples. Our methods have also successfully identified individuals within a mixture, a critical capability when considering touch samples.
It is important to note that proteomic analysis has some fundamental differences compared to conventional DNA genotyping. Most of these differences are caused by the process of transcription and translation. For DNA analysis, the absence of a marker can be as informative as its presence. This is because most forensic markers will have two copies in the genome and produce a concordant signal during analysis. Protein markers, on the other hand, will only be present if the gene is expressed in the appropriate tissue (e.g., skin). Even if the gene is expressed, there is no guarantee that both copies of the gene contributed equally to its expression. Further, variation in amino acid sequence between proteins can profoundly impact the detectability of a given protein variant using LC-MS, sometimes precluding the detection of one allele altogether. With these limitations in mind, Signature Science’s team has designed and developed statistical tools to calculate forensic match statistics that consider the unique nature of protein markers. We can calculate accurate random match probabilities (RMPs) and likelihood ratios (LRs) that reflect the specific nature of both protein markers and proteomic analysis. While we hope that the future holds a protein marker database analogous to CODIS, for now, these match statistics can be used to establish the likelihood that a protein profile from one sample matches another or that it matches to a specific suspect’s genome.
Conclusion:
We hope that our research and development efforts, alongside the work of other proteomic researchers, have established a strong foundation toward a future forensic capability. As the Proteos program concludes, we are eager to identify collaborators to further develop this emerging capability. Future efforts will help to inform the limitations of these methods and to identify the cases where protein sequencing could succeed when DNA analysis fails.
References
1. Parker, G. J. et al. Demonstration of Protein-Based Human Identification Using the Hair Shaft Proteome. PLoS ONE 11, e0160653 (2016).
- Stanciu, C. E., Philpott, M. K., Kwon, Y. J., Bustamante, E. E. & Ehrhardt, C. J. Optical characterization of epidermal cells and their relationship to DNA recovery from touch samples. F1000Research (2015) doi:10.12688/f1000research.7385.1.
- LeSassier, D. S. et al. Artificial fingerprints for cross-comparison of forensic DNA and protein recovery methods. PLOS ONE 14, e0223170 (2019).
- Schulte, K. Q. et al. Fractionation of DNA and protein from individual latent fingerprints for forensic analysis. Forensic Science International: Genetics 50, 102405 (2021).
Acknowledgements
This research is based upon work supported in part by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via contract number 2018-18041000003. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA,
or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein.
About the Author
Curt Hewitt, Ph.D. is one of the Principal Investigators leading Signature Science’s research and development efforts supporting IARPA’s Proteos program. He also leads applied research initiatives for Signature Science’s Center for Advanced Genomics. Beyond proteogenomics, Dr. Hewitt is currently focused on projects in synthetic biology, to include microbial engineering and coronavirus research, and human and microbial forensics.
For more information, please contact chewitt@signaturescience.com.
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!